9.1 The Genetic Code
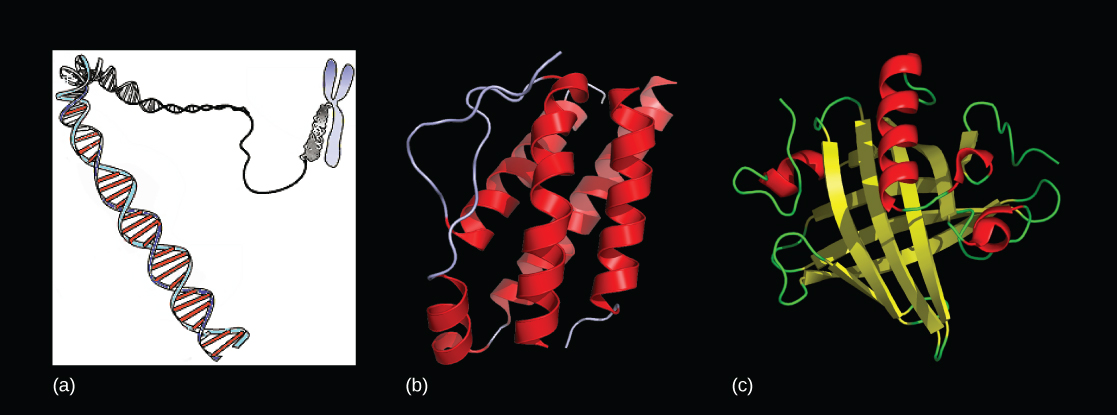
Since the rediscovery of Mendel’s work in 1900, the definition of the gene has progressed from an abstract unit of heredity to a tangible molecular entity capable of replication, expression, and mutation (Figure 9.1). Genes are composed of DNA and are linearly arranged on chromosomes. Genes specify the sequences of amino acids, which are the building blocks of proteins. In turn, proteins are responsible for orchestrating nearly every function of the cell. Both genes and the proteins they encode are absolutely essential to life as we know it.
Learning Objectives
By the end of this section, you will be able to:
- Explain the “central dogma” of DNA-protein synthesis
- Describe the genetic code and how the nucleotide sequence prescribes the amino acid and the protein sequence
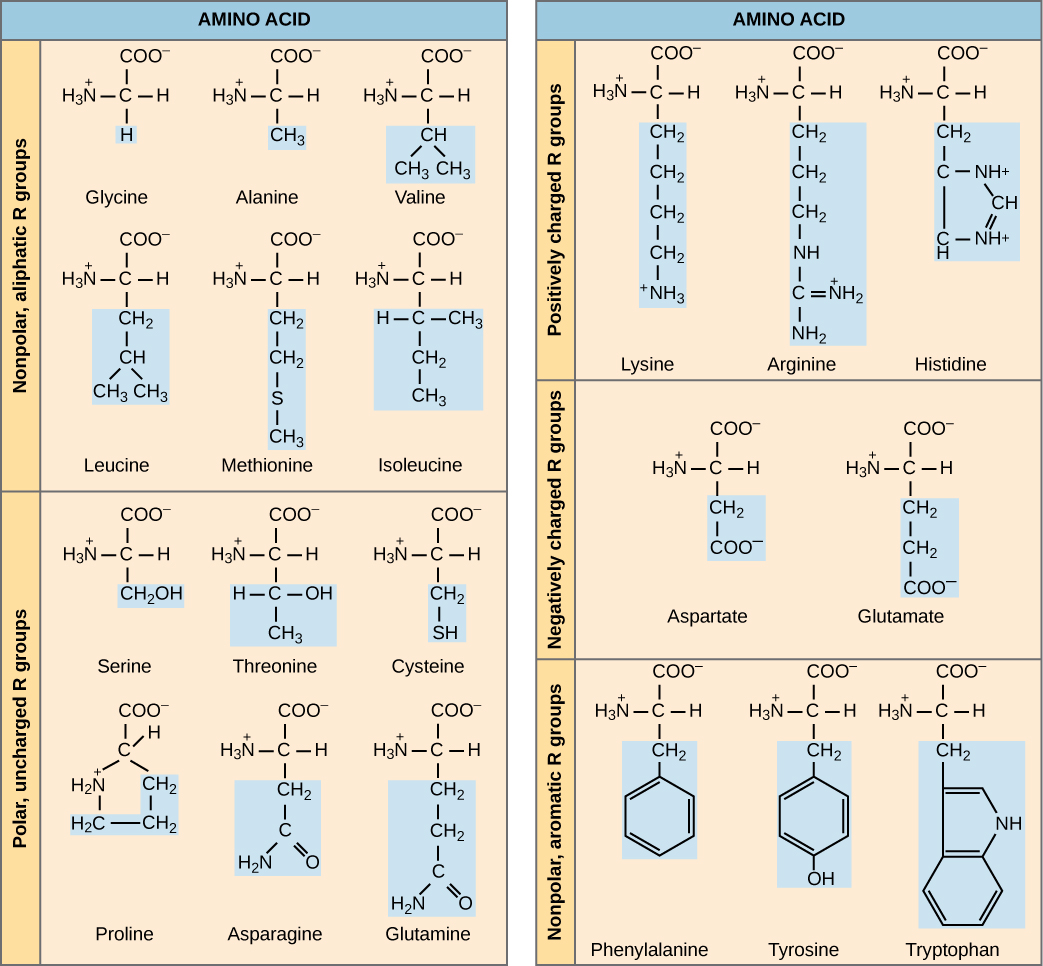
The cellular process of transcription generates messenger RNA (mRNA), a mobile molecular copy of one or more genes with an alphabet of A, C, G, and uracil (U). Translation of the mRNA template on ribosomes converts nucleotide-based genetic information into a protein product. That is the central dogma of DNA-protein synthesis. Protein sequences consist of 20 commonly occurring amino acids; therefore, it can be said that the protein alphabet consists of 20 “letters” (Figure 9.2). Different amino acids have different chemistries (such as acidic versus basic, or polar and nonpolar) and different structural constraints. Variation in amino acid sequence is responsible for the enormous variation in protein structure and function.
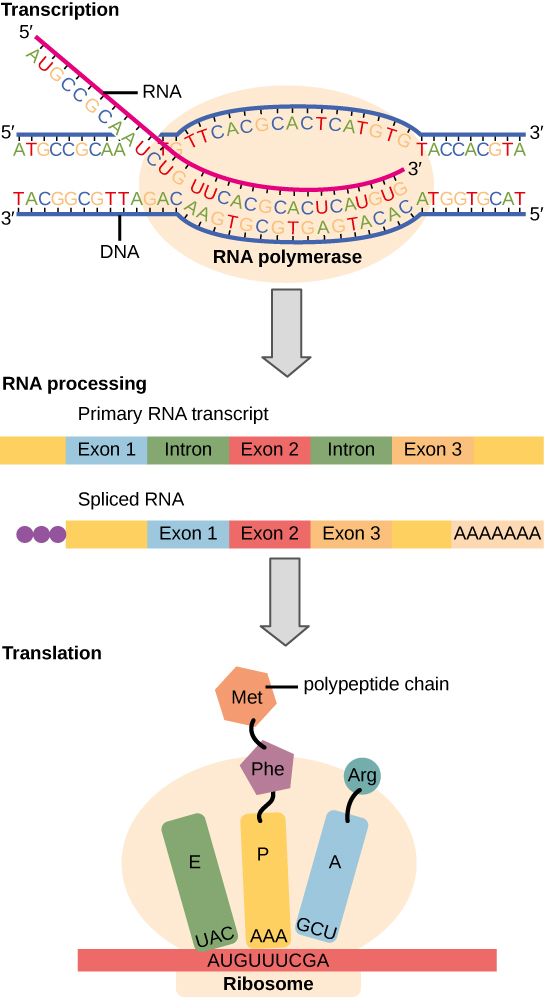
The Central Dogma: DNA Encodes RNA; RNA Encodes Protein
The flow of genetic information in cells from DNA to mRNA to protein is described by the central dogma (Figure 9.3), which states that genes specify the sequence of mRNAs, which in turn specify the sequence of amino acids making up all proteins. The decoding of one molecule to another is performed by specific proteins and RNAs. Because the information stored in DNA is so central to cellular function, it makes intuitive sense that the cell would make mRNA copies of this information for protein synthesis, while keeping the DNA itself intact and protected. The copying of DNA to RNA is relatively straightforward, with one nucleotide being added to the mRNA strand for every nucleotide read in the DNA strand. The translation to protein is a bit more complex because three mRNA nucleotides correspond to one amino acid in the polypeptide sequence. However, the translation to protein is still systematic and colinear, such that nucleotides 1 to 3 correspond to amino acid 1, nucleotides 4 to 6 correspond to amino acid 2, and so on.
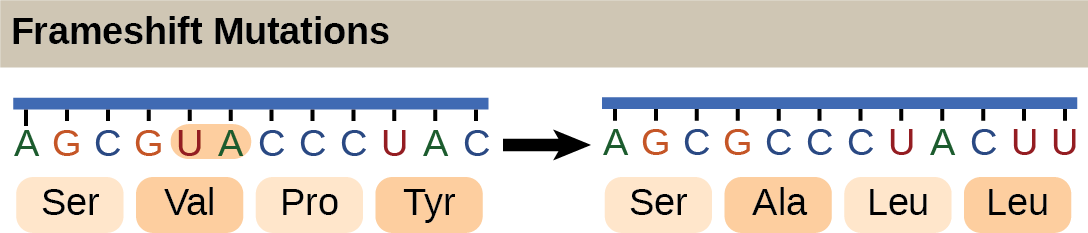
The Genetic Code Is Degenerate and Universal
Each amino acid is defined by a three-nucleotide sequence called the triplet codon. Given the different numbers of “letters” in the mRNA and protein “alphabets,” scientists theorized that single amino acids must be represented by combinations of nucleotides. Nucleotide doublets would not be sufficient to specify every amino acid because there are only 16 possible two-nucleotide combinations (42). In contrast, there are 64 possible nucleotide triplets (43), which is far more than the number of amino acids. Scientists theorized that amino acids were encoded by nucleotide triplets and that the genetic code was “degenerate.” In other words, a given amino acid could be encoded by more than one nucleotide triplet. This was later confirmed experimentally: Francis Crick and Sydney Brenner used the chemical mutagen proflavin to insert one, two, or three nucleotides into the gene of a virus. When one or two nucleotides were inserted, the normal proteins were not produced. When three nucleotides were inserted, the protein was synthesized and functional. This demonstrated that the amino acids must be specified by groups of three nucleotides. These nucleotide triplets are called codons. The insertion of one or two nucleotides completely changed the triplet reading frame, thereby altering the message for every subsequent amino acid (Figure 9.4). Though insertion of three nucleotides caused an extra amino acid to be inserted during translation, the integrity of the rest of the protein was maintained. Scientists painstakingly solved the genetic code by translating synthetic mRNAs in vitro and sequencing the proteins they specified (Figure 9.5).
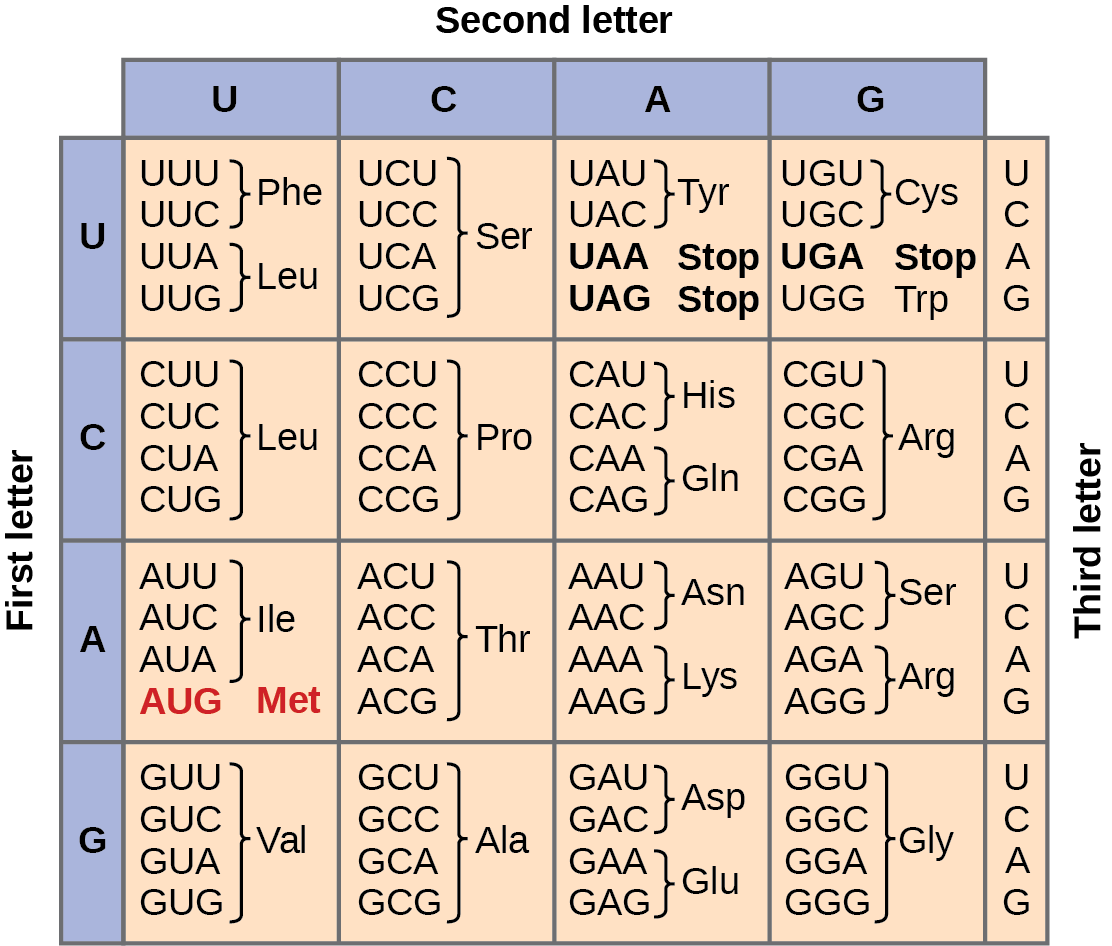
In addition to codons that instruct the addition of a specific amino acid to a polypeptide chain, three of the 64 codons terminate protein synthesis and release the polypeptide from the translation machinery. These triplets are called nonsense codons, or stop codons. Another codon, AUG, also has a special function. In addition to specifying the amino acid methionine, it also serves as the start codon to initiate translation. The reading frame for translation is set by the AUG start codon near the 5′ end of the mRNA. Following the start codon, the mRNA is read in groups of three until a stop codon is encountered.
The arrangement of the coding table reveals the structure of the code. There are sixteen “blocks” of codons, each specified by the first and second nucleotides of the codons within the block, e.g., the “AC*” block that corresponds to the amino acid threonine (Thr). Some blocks are divided into a pyrimidine half, in which the codon ends with U or C, and a purine half, in which the codon ends with A or G. Some amino acids get a whole block of four codons, like alanine (Ala), threonine (Thr) and proline (Pro). Some get the pyrimidine half of their block, like histidine (His) and asparagine (Asn). Others get the purine half of their block, like glutamate (Glu) and lysine (Lys). Note that some amino acids get a block and a half-block for a total of six codons.
The specification of a single amino acid by multiple similar codons is called “degeneracy.” Degeneracy is believed to be a cellular mechanism to reduce the negative impact of random mutations. Codons that specify the same amino acid typically only differ by one nucleotide. In addition, amino acids with chemically similar side chains are encoded by similar codons. For example, aspartate (Asp) and glutamate (Glu), which occupy the GA* block, are both negatively charged. This nuance of the genetic code ensures that a single-nucleotide substitution mutation might specify the same amino acid but have no effect or specify a similar amino acid, preventing the protein from being rendered completely nonfunctional.
The genetic code is nearly universal. With a few minor exceptions, virtually all species use the same genetic code for protein synthesis. Conservation of codons means that a purified mRNA encoding the globin protein in horses could be transferred to a tulip cell, and the tulip would synthesize horse globin. That there is only one genetic code is powerful evidence that all of life on Earth shares a common origin, especially considering that there are about 1084 possible combinations of 20 amino acids and 64 triplet codons.
Transcribe a gene and translate it to protein using complementary pairing and the genetic code at this site.
Which Has More DNA: A Kiwi or a Strawberry?

Question: Would a kiwi and strawberry that are approximately the same size (Figure 9.6) also have approximately the same amount of DNA?
Background: Genes are carried on chromosomes and are made of DNA. All mammals are diploid, meaning they have two copies of each chromosome. However, not all plants are diploid. The common strawberry is octoploid (8n) and the cultivated kiwi is hexaploid (6n). Research the total number of chromosomes in the cells of each of these fruits and think about how this might correspond to the amount of DNA in these fruits’ cell nuclei. What other factors might contribute to the total amount of DNA in a single fruit? Read about the technique of DNA isolation to understand how each step in the isolation protocol helps liberate and precipitate DNA.
Hypothesis: Hypothesize whether you would be able to detect a difference in DNA quantity from similarly sized strawberries and kiwis. Which fruit do you think would yield more DNA?
Test your hypothesis: Isolate the DNA from a strawberry and a kiwi that are similarly sized. Perform the experiment in at least triplicate for each fruit
- Prepare a bottle of DNA extraction buffer from 900 mL water, 50 mL dish detergent, and two teaspoons of table salt. Mix by inversion (cap it and turn it upside down a few times).
- Grind a strawberry and a kiwi by hand in a plastic bag, or using a mortar and pestle, or with a metal bowl and the end of a blunt instrument. Grind for at least two minutes per fruit.
- Add 10 mL of the DNA extraction buffer to each fruit, and mix well for at least one minute.
- Remove cellular debris by filtering each fruit mixture through cheesecloth or porous cloth and into a funnel placed in a test tube or an appropriate container.
- Pour ice-cold ethanol or isopropanol (rubbing alcohol) into the test tube. You should observe white, precipitated DNA.
- Gather the DNA from each fruit by winding it around separate glass rods.
Record your observations: Because you are not quantitatively measuring DNA volume, you can record for each trial whether the two fruits produced the same or different amounts of DNA as observed by eye. If one or the other fruit produced noticeably more DNA, record this as well. Determine whether your observations are consistent with several pieces of each fruit.
Analyze your data: Did you notice an obvious difference in the amount of DNA produced by each fruit? Were your results reproducible?
Draw a conclusion: Given what you know about the number of chromosomes in each fruit, can you conclude that chromosome number necessarily correlates to DNA amount? Can you identify any drawbacks to this procedure? If you had access to a laboratory, how could you standardize your comparison and make it more quantitative?
Section Summary
The genetic code refers to the DNA alphabet (A, T, C, G), the RNA alphabet (A, U, C, G), and the polypeptide alphabet (20 amino acids). The central dogma describes the flow of genetic information in the cell from genes to mRNA to proteins. Genes are used to make mRNA by the process of transcription; mRNA is used to synthesize proteins by the process of translation. The genetic code is degenerate because 64 triplet codons in mRNA specify only 20 amino acids and three nonsense codons. Most amino acids have several similar codons. Almost every species on the planet uses the same genetic code.
Exercises
Glossary
central dogma: states that genes specify the sequence of mRNAs, which in turn specify the sequence of proteins
codon: three consecutive nucleotides in mRNA that specify the insertion of an amino acid or the release of a polypeptide chain during translation
colinear: in terms of RNA and protein, three “units” of RNA (nucleotides) specify one “unit” of protein (amino acid) in a consecutive fashion
degeneracy (of the genetic code): describes that a given amino acid can be encoded by more than one nucleotide triplet; the code is degenerate, but not ambiguous
nonsense codon: one of the three mRNA codons that specifies termination of translation
reading frame: sequence of triplet codons in mRNA that specify a particular protein; a ribosome shift of one or two nucleotides in either direction completely abolishes synthesis of that protein
Media Attributions
- Figure 9.1 “chromosome”: National Human Genome Research Institute; “interleukin-2”: Ramin Herati/Created from PDB 1M47 and rendered with Pymol; “alpha-2u-globulin”: Darren Logan/rendered with AISMIG
- Figure 9.5 modification of work by NIH
- Figure 9.6 “kiwi”: Kelbv/Flickr; “strawberry”: Alisdair McDiarmid

